Lubomir Vezenkov
ASSOCIATE Professor, National Graduate School of Chemistry of Montpellier
Lubomir Vezenkov was born in Bulgaria, where he performed his studies until he arrived in Montpellier in 2005 for his masters in the ENSCM engineering school.
Then, he performed a co-tutoring PhD between the universities of Montpellier and Naples, which he graduated from in 2011. During this period, he helped develop some potent cell penetrating foldamers that were used to vectorize pepststin and provide selective anti-cancer activity.
Afterwards, he did a two-years-long post-doctoral internship in the prof. Robert Young group in Simon Fraser University, Vancouver, where he took part in the development of one the first specific autophagy inhibitors.
Then, he moved back to Montpellier, and in 2016, he received an associate professor position at ENSCM in team 9 of the IBMM research institute.
Currently, he is working in the fields of peptide and medicinal chemistry as well as in the exciting domain of chemical biology. Recently he has been interested in the development of covalent-irreversible and covalent-reversible protease inhibitors. These covalent modifiers are used to block or activate certain microtubulin post-translational modifications. Such compounds can help better understand the polarization of neurons and can be used as therapeutics for neurodegenerative disease.
lubomir.vezenkov@enscm.fr
+33 448792185
5 major publications :
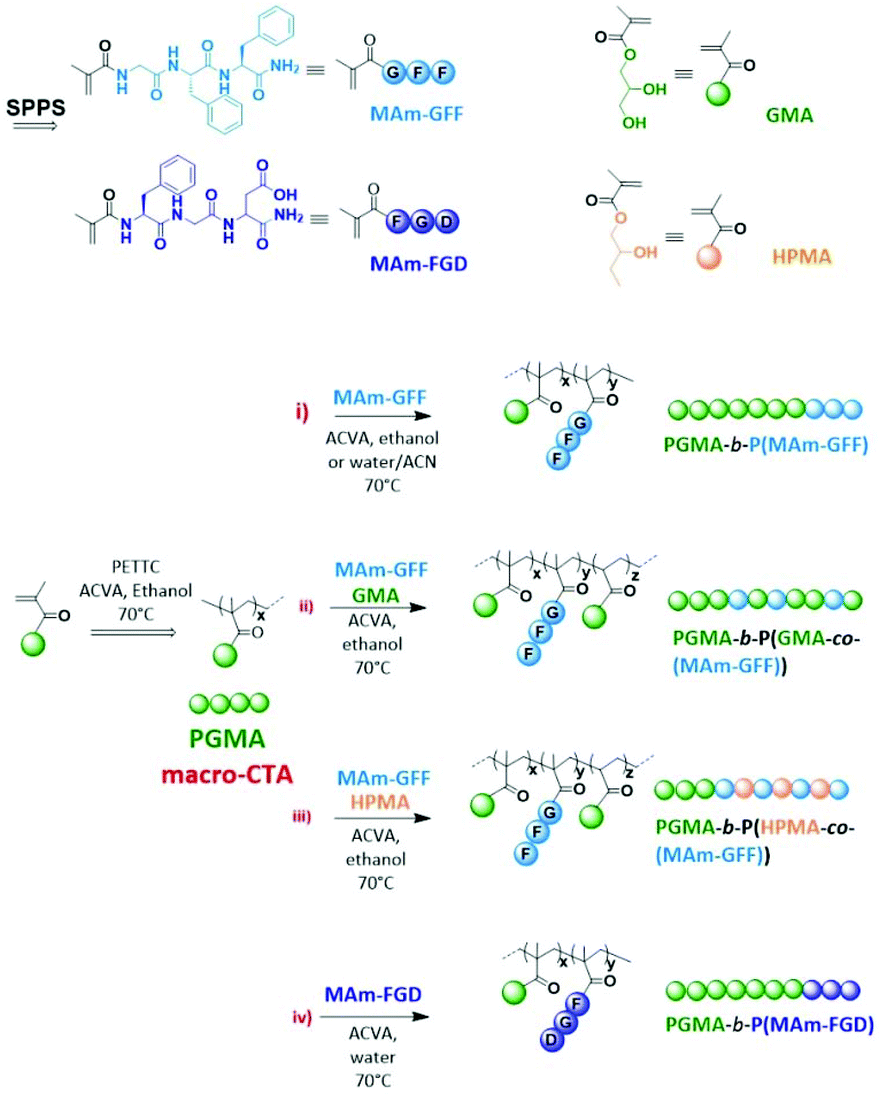
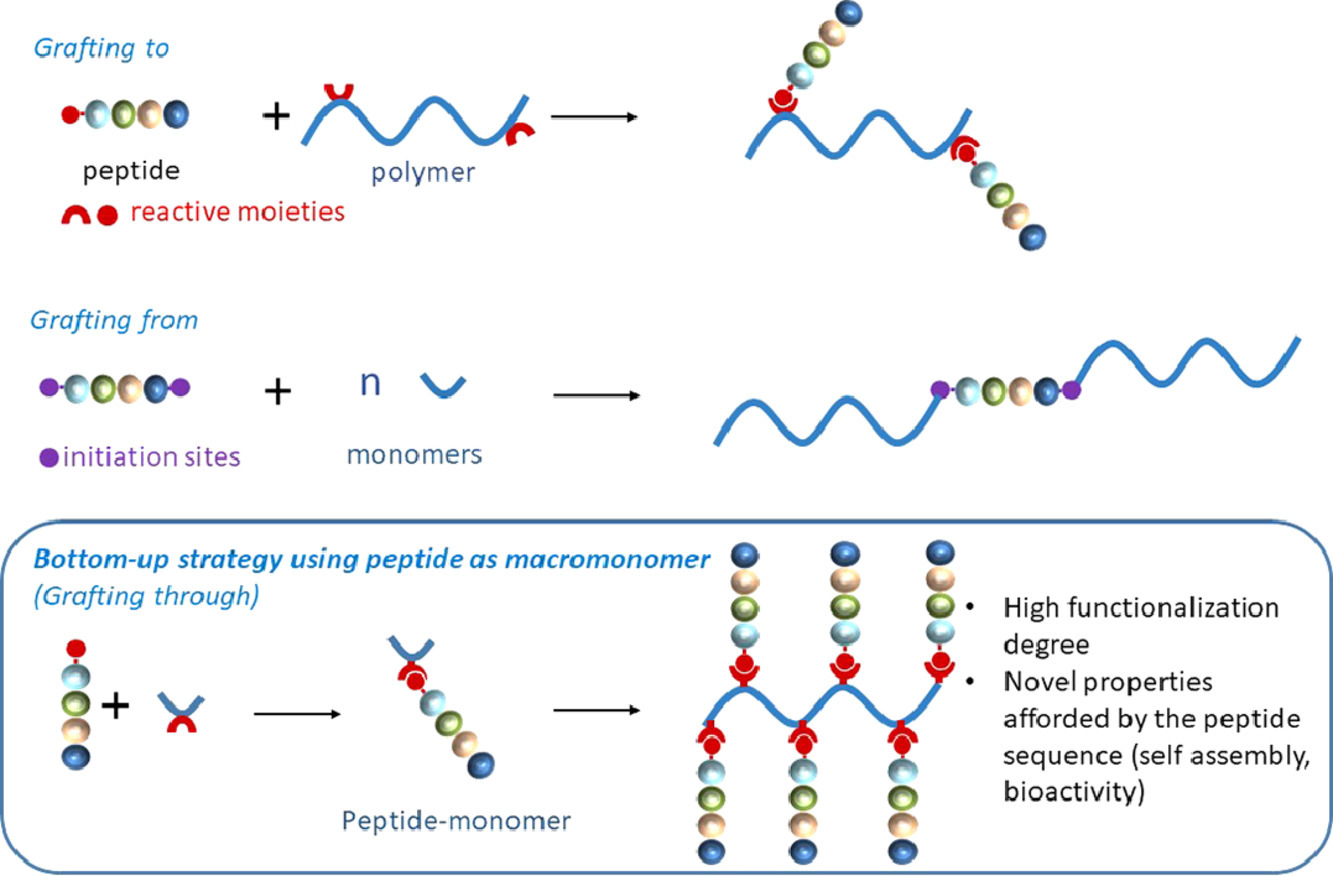
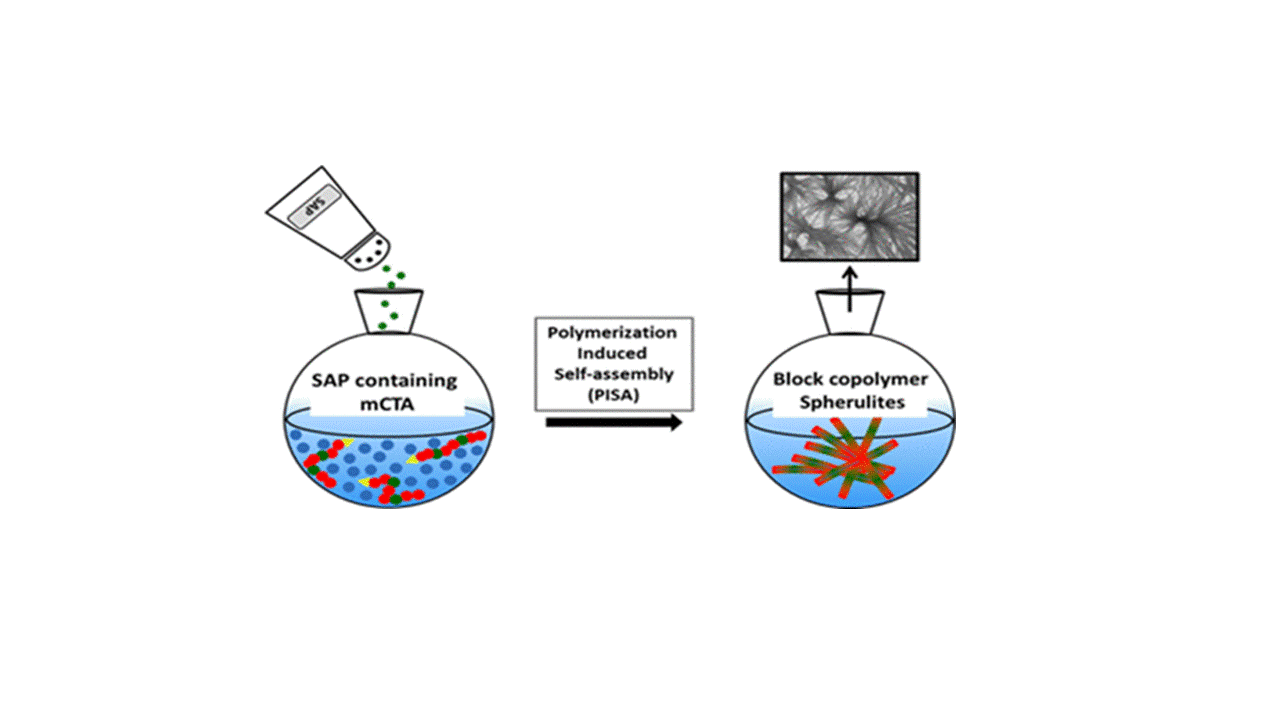
Dao T, Vezenkov L, Subra G, Amblard M, In M, Le Meins J-F, Aubrit F, Moradi M-A, Ladmiral V, Semsarilar M; Self-Assembling Peptide—Polymer Nano-Objects via Polymerization-Induced Self-Assembly, Macromolecules 2020, 53, 16, 7034–7043. https://doi.org/10.1021/acs.macromol.0c01260
Bosc D, Vezenkov L, Bortnik S, An J, Xu J, Choutka C, Hannigan AM, Kovacic S, Loo S, Clark PGK, Chen G, Guay-Ross RN, Yang K, Dragowska WH, Zhang F, Go NE, Leung A, Honson NS, Pfeifer TA, Gleave M, Bally M, Jones SJ, Gorski SM, Young RN; A new quinoline-based chemical probe inhibits the autophagy-related cysteine protease ATG4B, Sci Rep., 2018 Aug 3;8(1):11653. doi: 10.1038/s41598-018-29900-x.
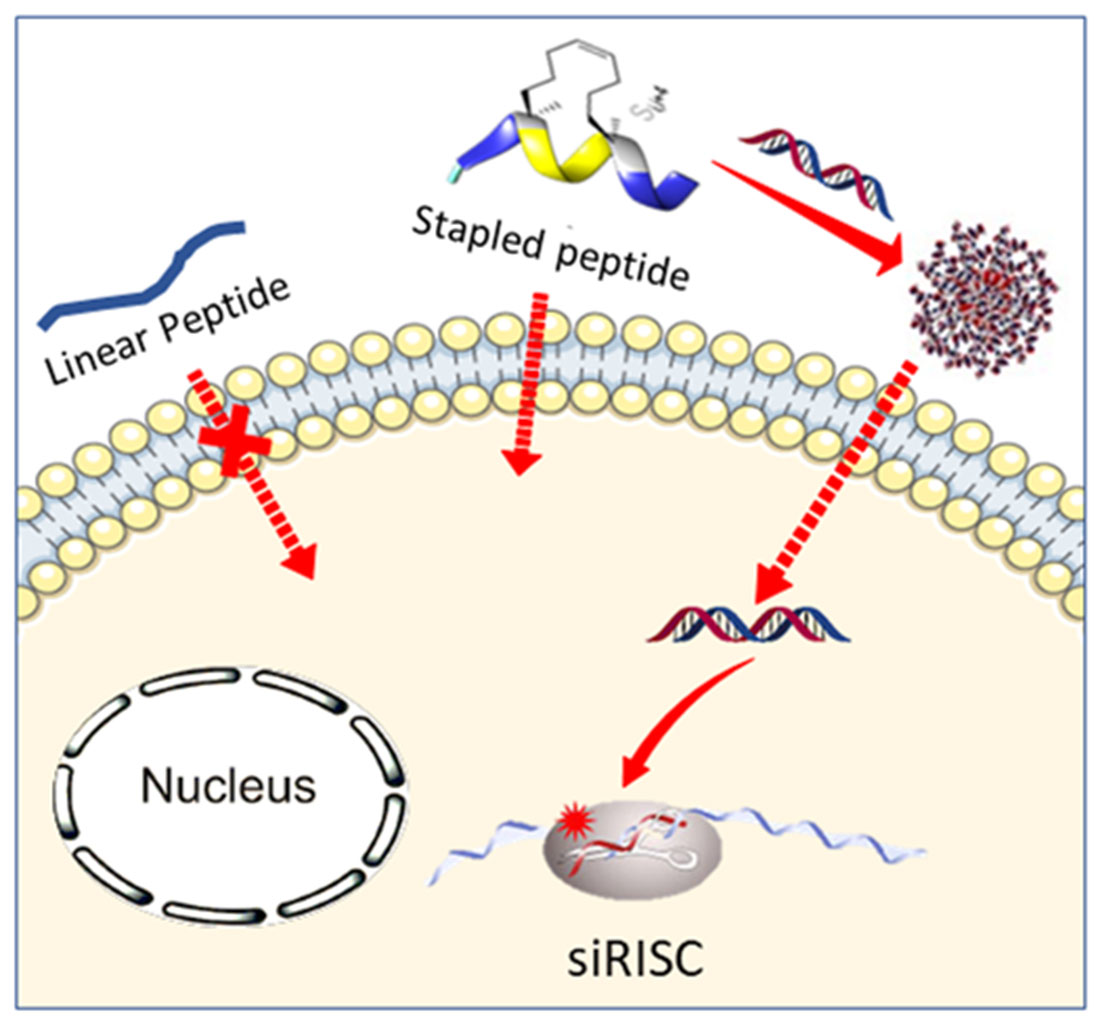
Vezenkov L, Martin V, Bettache N, Simon M, Messerschmitt A, Legrand B, Bantignies JL, Subra G, Maynadier M, Bellet V, Garcia M, Martinez J, Amblard M; Ribbon-like Foldamers for Cellular Uptake and Drug Delivery, Chembiochem, 2017 Nov 2;18(21):2110-2114 https://doi.org/10.1002/cbic.201700455
Vezenkov L, Maynadier M, Amblard M, Martin V, Gandreuil C, Vaillant O, Gary-Bobo M, Basile I, Hernandez JF, Garcia M, Martinez J; Dipeptide mimic oligomer transporter mediates intracellular delivery of Cathepsin D inhibitors: a potential target for cancer therapy. Journal of Controlled Release, 2013 Oct 28;171(2):251-7. https://doi.org/10.1016/j.jconrel.2013.07.017Get rights and content
Vezenkov L, Maynadier M, Hernandez JF, Averlant-Petit M, Fabre O, Garcia M, Martinez J, Amblard M; Noncationic Dipeptide Mimic Oligomers As Cell Penetrating Nonpeptides(CPNP), Bioconjugate Chem. 2010, Sept 21(10), 1850–1854. https://doi.org/10.1021/bc1002086